Generalizable, Reproducible, and Neuroscientifically Interpretable Imaging Biomarkers for Alzheimer's Disease
Dan Jin, Bo Zhou, Ying Han, Jiaji Ren, Tong Han, Bing Liu, Jie Lu, Chengyuan Song, Pan Wang, Dawei Wang, Jian Xu, Zhengyi Yang, Hongxiang Yao, Chunshui Yu, Kun Zhao, Max Wintermark, Nianming Zuo, Xinqing Zhang, Yuying Zhou, Xi Zhang, Tianzi Jiang, Qing Wang,* and Yong Liu*
Abstract:
Precision medicine for Alzheimer's disease (AD) necessitates the development of personalized, reproducible, and neuroscientifically interpretable biomarkers, yet despite remarkable advances, few such biomarkers are available. Also, a comprehensive evaluation of the neurobiological basis and generalizability of the end‐to‐end machine learning system should be given the highest priority. For this reason, a deep learning model (3D attention network, 3DAN) that can simultaneously capture candidate imaging biomarkers with an attention mechanism module and advance the diagnosis of AD based on structural magnetic resonance imaging is proposed. The generalizability and reproducibility are evaluated using cross‐validation on in‐house, multicenter (n = 716), and public (n = 1116) databases with an accuracy up to 92%. Significant associations between the classification output and clinical characteristics of AD and mild cognitive impairment (MCI, a middle stage of dementia) groups provide solid neurobiological support for the 3DAN model. The effectiveness of the 3DAN model is further validated by its good performance in predicting the MCI subjects who progress to AD with an accuracy of 72%. Collectively, the findings highlight the potential for structural brain imaging to provide a generalizable, and neuroscientifically interpretable imaging biomarker that can support clinicians in the early diagnosis of AD.
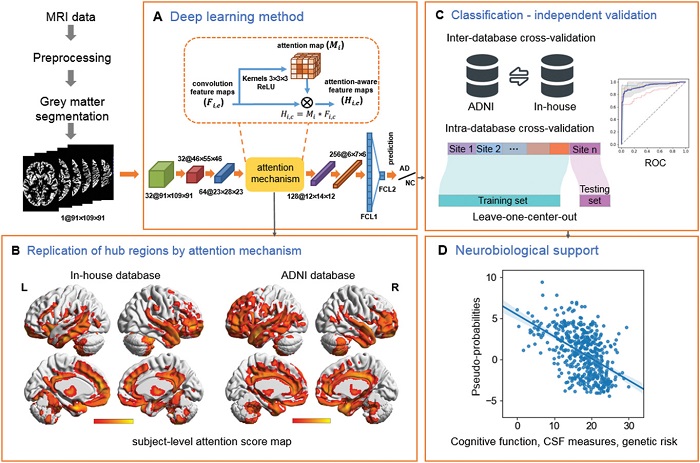 | Figure. Schematic of the data analysis pipeline. A) The architecture of the 3D attention network (3DAN). In the attention mechanism module, each voxel i of the H × W × D‐dimensional feature maps F i ,c was weighted by the H × W × D‐dimensional attention map Mi . The trainable attention map Mi was independent of the channel of the features and was only related to the spatial position. B) The attention score map (left: in‐house database, right: ADNI database) was generated by the attention mechanism module of the 3DAN model, indicating the discriminative power of various brain regions for AD diagnosis. C) To test the robustness and generalizability of the 3DAN model, cross validations were performed using two completely independent databases (an in‐house database and the ADNI database) (Details can be found in Table 1). D) Investigation of the association between the classification output and clinical measures [that is the cognitive function measured by Mini‐Mental State Examination (MMSE), CSF beta‐amyloid (Aβ ), CSF tau, and polygenic risk scores (PGRS)] in the AD and MCI groups. |
